| CAT No | Product | Size | Price |
|---|---|---|---|
| PB72.10-01 | VeriFi® Library Amplification Mix | 50 x 50 μL Reactions | Contact us |
| PB72.10-05 | VeriFi® Library Amplification Mix | 250 x 50 μL Reactions | Contact us |
To view your prices, please log in or register with your quote code.
Additional Information
VeriFi® Library Amplification Mix is designed as a tool for researchers performing next-generation sequencing (NGS) studies. An important part of the PCR-based NGS workflow is the amplification step, where a DNA or cDNA library (in the case of RNA-Seq studies) has been modified to contain the appropriate adapters for the intended sequencing platform and needs to be amplified before proceeding to the sequencing step.
To ensure the accuracy of the final sequencing data, proofreading polymerases used in the amplification step of NGS workflows must have key characteristics, such as:
- High accuracy, to minimize PCR errors in nucleotide incorporation.
- Strong multiplexing capability, to amplify different fragment lengths in a library.
- Low sequence-dependent bias, especially when dealing with complex sequences (GC/AT-rich, or repetitive).
Additional features may also help improve the performance of the library amplification mix, depending on the type of NGS study being conducted. VeriFi® Library Amplification Mix is designed with these features in mind. It is powered by our market-leading VeriFi® polymerase, with AptaLock™ hot start technology, in a new 2x mix format. This mix contains reaction buffer, dNTPs, Mg2+, and enhancers that help minimize GC bias and enable reliable library amplification for Illumina sequencing platforms. Amplified libraries can be easily quantified using the Blue NGSBIO Library Quantification Kit for Illumina before sequencing.
Better Quality NGS Data
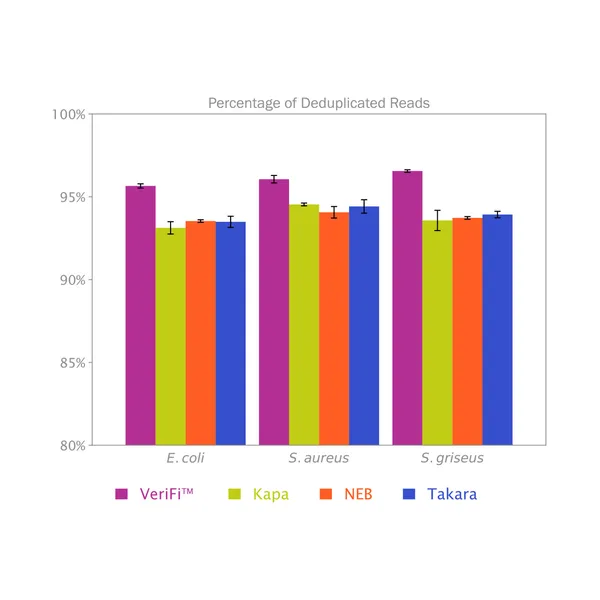
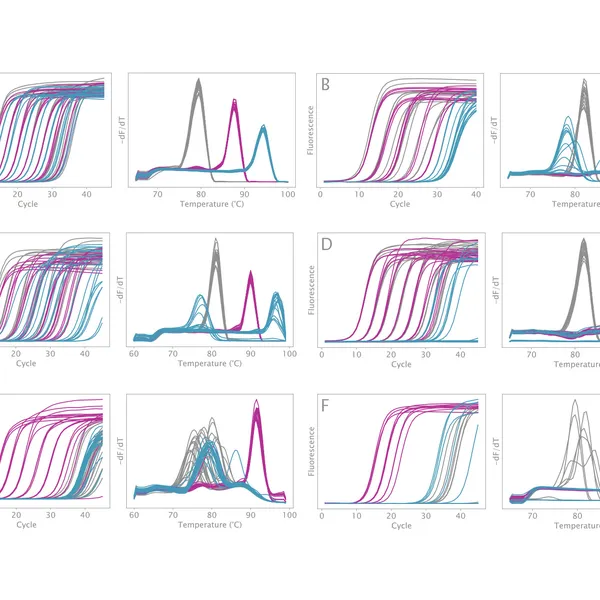
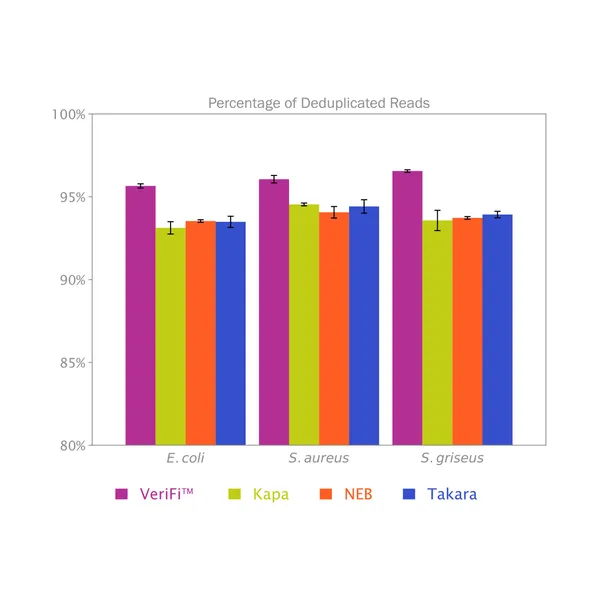
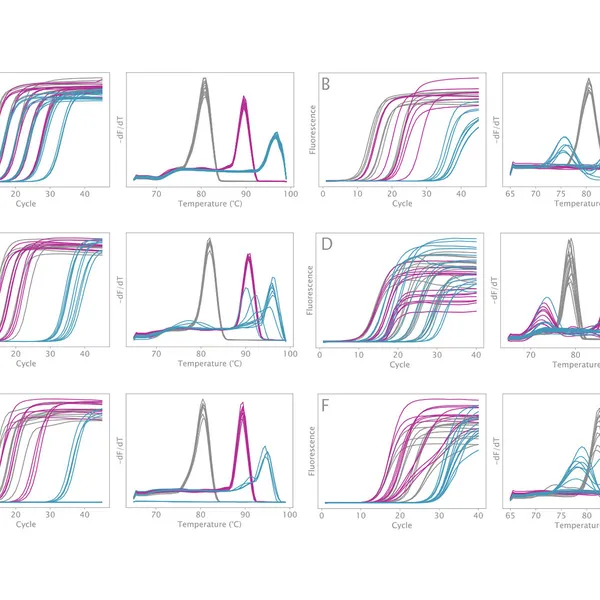
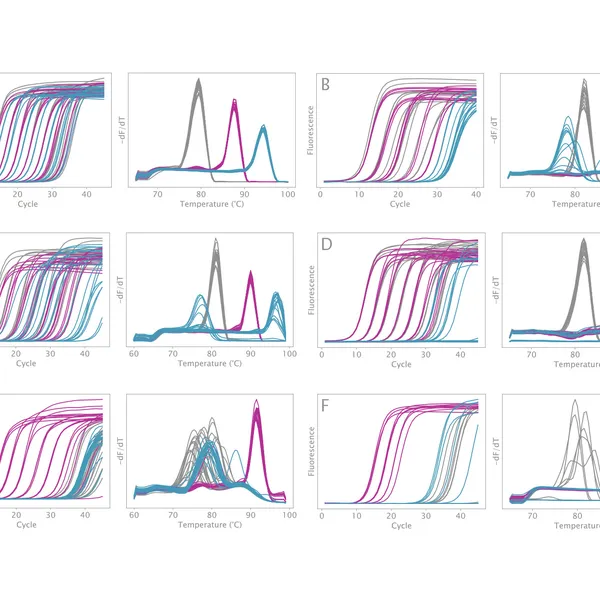
VeriFi® Library Amplification Mix has been externally validated using Illumina-based sequencing in a blind comparison with three leading competitors on the market. VeriFi® Library Amplification Mix consistently delivers 2% more uniquely mapped reads than competitors for three different microbial genomes. Depending on the sequencing platform used, this small percentage can translate to 80,000 to 100,000,000 more unique reads. This results in greater target sequence coverage and more precise quantitative information, making sequencing experiments more cost-effective and resulting data more informative.
Reducing GC Bias in NGS Library Amplification
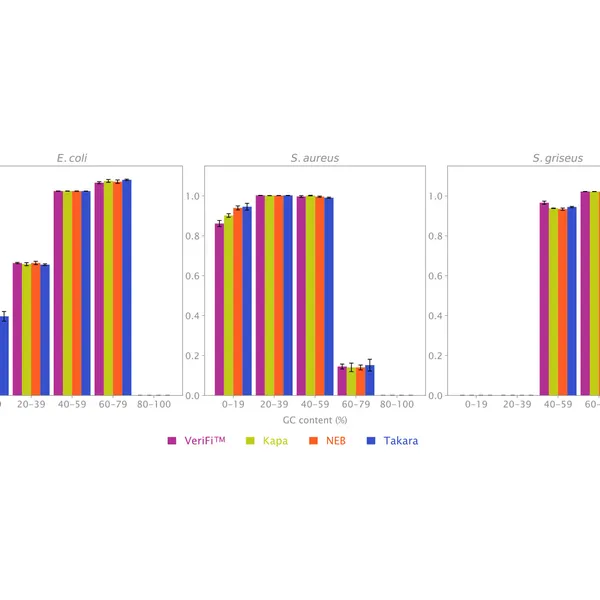
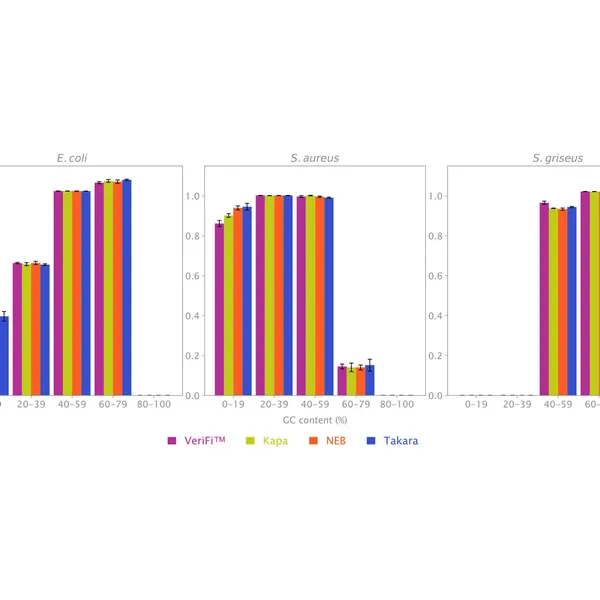
The new buffer formulation makes VeriFi® Library Amplification Mix ideal for complex samples. Our research team has challenged this enzyme in PCRs on numerous difficult templates, demonstrating that there is minimal bias in amplification across extremes of GC content from 30% – 70%. With consistently superior performance to all major competitors, these results ensure digested performance of the mix regardless of target sequence, allowing you confidence in conducting costly and time-consuming NGS experiments. Paired with the increased number of unique reads, discussed above, this feature makes VeriFi® Library Amplification Mix ideal for metagenomic analyses.
AptaLock™ Hot Start Technology
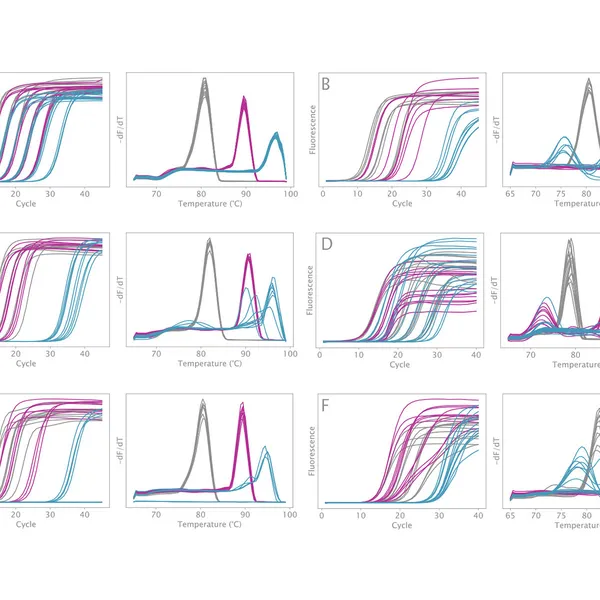
VeriFi® Library Amplification Mix is enhanced by PCRBIO's innovative AptaLock™ Hot Start technology, a distinct aptamer-like molecule that temporarily inhibits both the 3'-5' exonuclease activity and 5'-3' polymerase activity of the enzyme at ambient temperature. This unique hot start molecule prevents primer-dimer formation and non-specific amplification to maximize PCR sensitivity and specificity. This ensures that amplification reactions can be set up at room temperature.
References
- Chen, Y.C., Liu, T., Yu, C.H., Chiang, T.Y., Hwang, C.C., The impact of GC bias in next-generation sequencing data on De Novo</i> Genome Assembly. PLOS ONE 8(4): e62856. ref=”https://doi.org/10.1371/journal.pone.0062856
- Sims, D., Sudbery, I., Ilott, N. et al. Sequencing depth and coverage: key considerations in genomic analysis. Nat Rev Genet 15, 121–132 (2014). ref=”https://doi.org/10.1038/nrg3642
Applications
- NGS Library Amplification
- Whole Genome Sequencing
- RNA-Seq
- High-Throughput and Multiplex PCR
- Targeted Sequencing of GC/AT-Rich Regions
- Metagenomic Studies
Technical Specifications
VeriFi® Library Amplification Mix
| Component | 50 Reactions | 250 Reactions |
|---|---|---|
| 2x VeriFi Library Amplification Mix | 1 x 1.25 mL | 5 x 1.25 mL |
VeriFi® Library Amplification Mix
| Component | 50 Reactions | 250 Reactions |
|---|---|---|
| 2x VeriFi Library Amplification Mix | 1 x 1.25 mL | 5 x 1.25 mL |
Reaction Information
| Reaction Volume | Storage | |||
|---|---|---|---|---|
| 50 μL | Upon receipt, the product should be stored at a temperature between -30 and -20 °C. If stored correctly, the kit will maintain full activity until the specified expiry date. |
Documents
Product Brochure
Safety Data Sheet
Application Notes
Frequently Asked Questions (FAQs)
Can I perform multiplex PCR with VeriFi® Library Amplification Mix?
Yes. For most multiplex experiments, we recommend using VeriFi® Hot Start Polymerase & Mixes (product numbers PB10.45-PB10.47). However, VeriFi® Library Amplification Mix is also well-suited for this purpose, particularly when one or more multiplex targets contain GC-rich regions or have challenging or repetitive sequence regions.
Can I use VeriFi® Library Amplification Mix for bisulfite-treated DNA methylation studies/amplification?
No. Pfu polymerases can only amplify targets containing uracil following transformation, which reduces PCR accuracy.
Can I use VeriFi® Library Amplification Mix for standard PCR instead of library amplification?
Yes, please follow the relevant guidance in the product manual. VeriFi® Library Amplification Mix can be used in standard 3-step or 2-step cycling programs and is particularly suited for amplifying targets with very high or very low GC content.
Can VeriFi® Library Amplification Mix be used in single-cell NGS experiments?
VeriFi® Library Amplification Mix can be used for PCR-based enrichment of any type of DNA library.
Do I need to include a purification step between adaptor ligation and library amplification with VeriFi® Library Amplification Mix?
We recommend including a purification step after adaptor ligation and size selection. This removes any buffer incompatibilities and ensures that unligated adaptors are not amplified during the PCR library enrichment.
I am having difficulty amplifying a target with high GC content, what should I do?
Increase the denaturation temperature to 98-100 °C and increase the extension time.
My amplified library is of low quality, how can I improve it?
Increase denaturation temperature. Optimize annealing temperature and extension time. Optimize primer and template amounts in the reaction. Ensure the starting material is of high quality.
What is the maximum target length I can amplify with VeriFi® Library Amplification Mix?
We have successfully amplified up to a 12 kb fragment containing GC-rich regions. Longer targets can also be successfully tested.
What is the minimum sample amount I can use for successful library amplification with VeriFi® Library Amplification Mix?
Do not use less than 10 ng of template for NGS library amplification. A key factor in obtaining high-quality amplified libraries is limiting the number of PCR cycles used. Using higher cycle numbers will result in a greater risk of errors and bias caused by PCR.
What NGS sequencing platforms can I use VeriFi® Library Amplification Mix with?
VeriFi® Library Amplification Mix has been extensively tested and validated on Illumina sequencing platforms with Illumina P5 and P7 primers. The mix should also work well in amplifying libraries with different adapter sequences intended for other sequencing platforms (e.g., Pac Bio and Oxford Nanopore Technologies), but this should be verified.


Read more