| CAT Code | Product | Size | Price |
|---|---|---|---|
| PB20.32-01 | Clara® HRM Mix | 100 x 20 μL Reactions | Contact us |
| PB20.32-05 | Clara® HRM Mix | 500 x 20 μL Reactions | Contact us |
| PB20.32-20 | Clara® HRM Mix | 2000 x 20 μL Reactions | Contact us |
To see your price, please log in or register with your price quote code.
Additional Information
What is HRM?
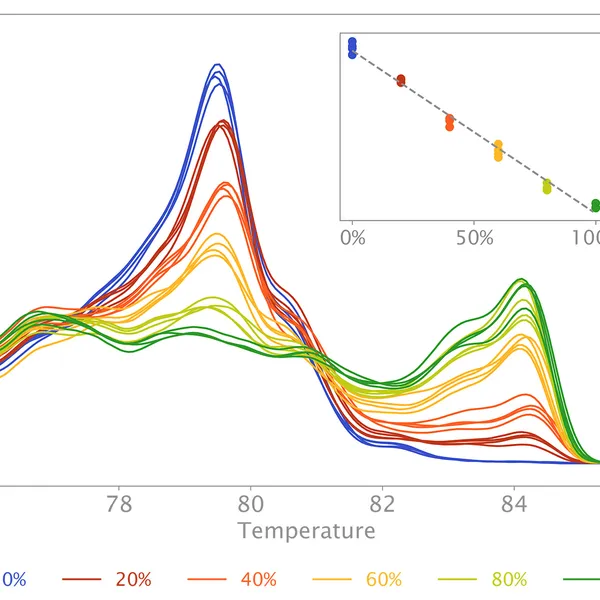
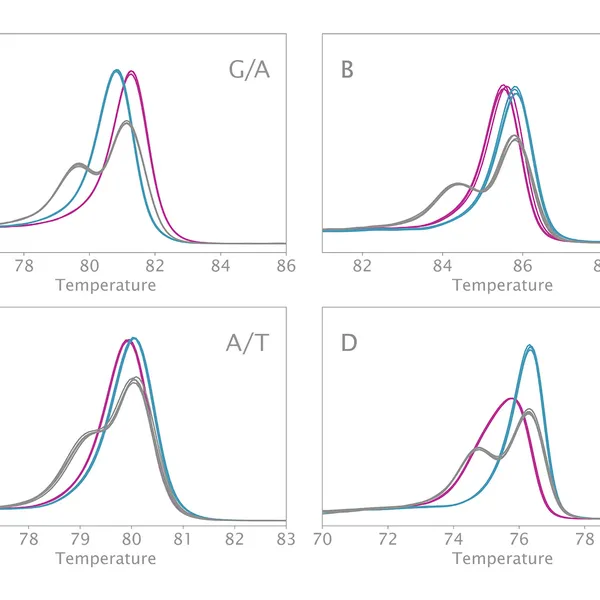
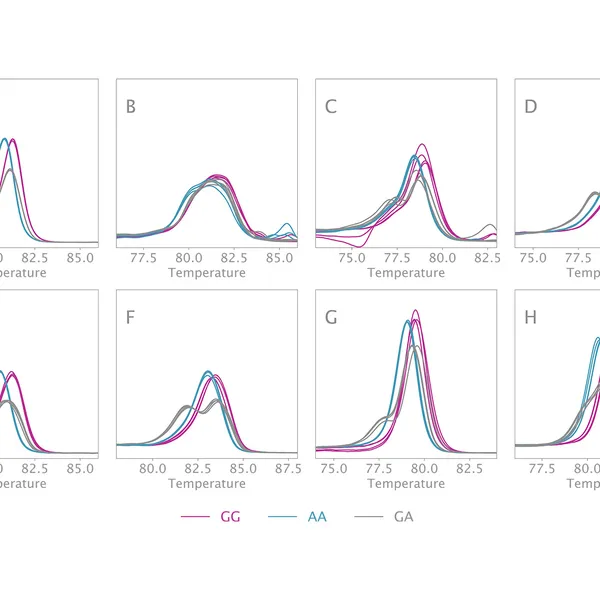
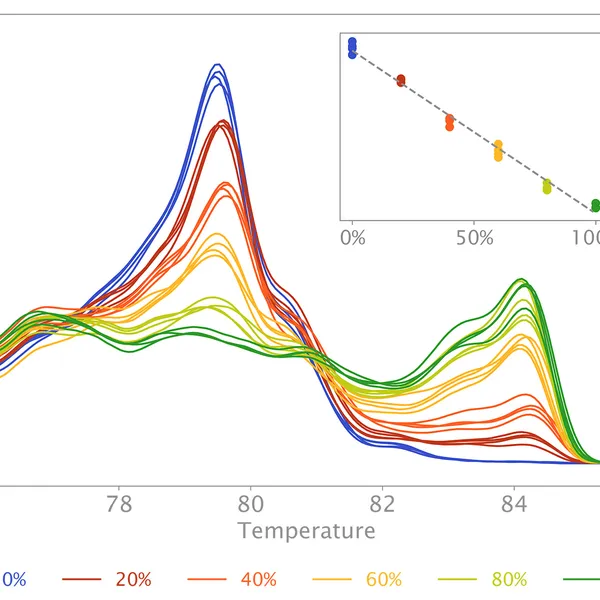
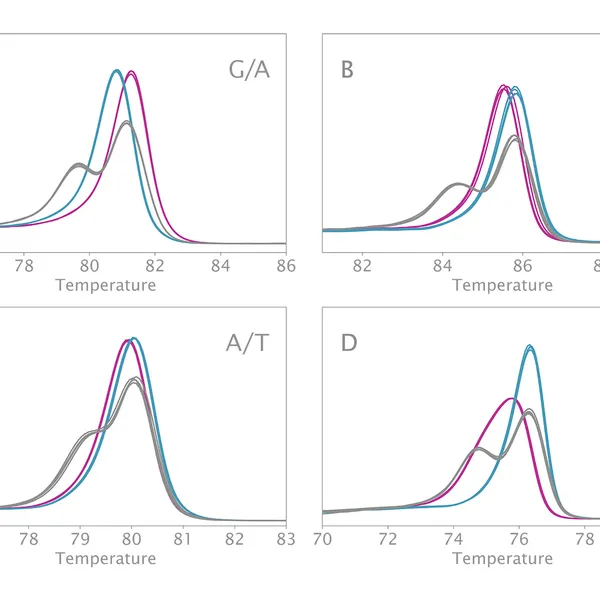
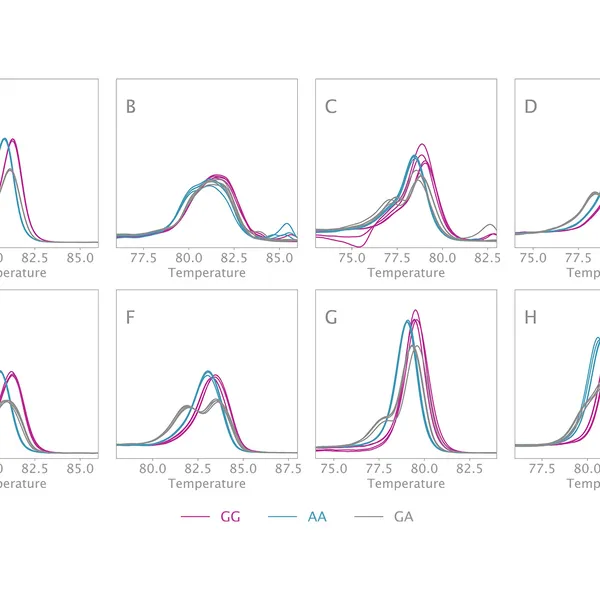
High Resolution Melting analysis (HRM) is a powerful technique used to identify variants, polymorphisms, and epigenetic differences in double-stranded DNA samples. This approach relies on analyzing the post-PCR-generated melting profiles of amplified targets in conjunction with third-generation DNA intercalating dyes designed to minimize polymerase inhibition (e.g., SYBR Green 2, EvaGreen). HRM makes use of differences in melting curve shape and melting temperature of DNA to differentiate between sequence variants of samples.
Typically, in post-PCR melting curve analysis with standard dye-based qPCR, fluorescence data are collected every 0.2-0.5 °C, whereas in HRM analysis, data are gathered at a much higher density, every 0.1 °C, resulting in significantly higher melting curve resolution. Thus, even the smallest differences (down to single base pair mutations, SNPs, and base-pair methylations) within the target sequences are recorded as shifted melt curves. HRM thereby provides a more efficient, cost-effective solution for SNP detection, allele discrimination, and target variant discovery.
Detectable Sequence Variations
Clara® HRM Mix can differentiate all SNP types. These types include:
- Type I: C to T and G to A
- Type II: C to A and G to T
- Type III: C to G
- Type IV: A to T
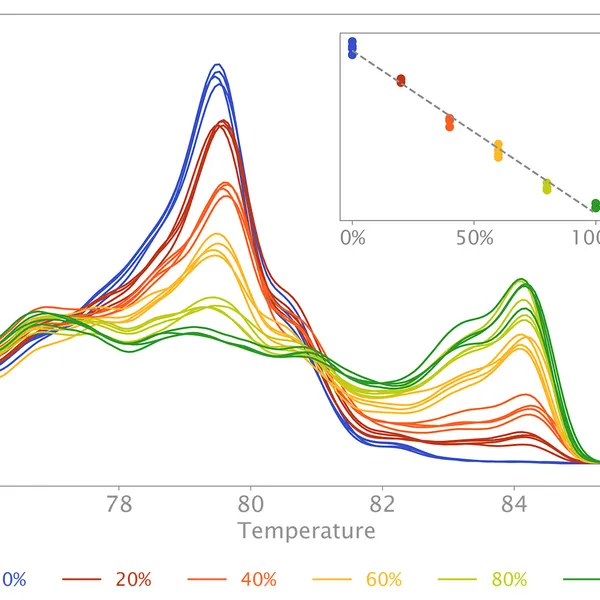
Additionally, the mix can also be used to quantify CpG methylation in a target sequence. In this case, methylated DNA will need to be bisulfite-treated before HRM qPCR with primers targeting CpG-containing regions. During bisulfite conversion, all unmethylated cytosine residues will be deaminated and thus converted to uracil residues, whereas all methylated cytosine residues will remain unchanged. HRM analysis on bisulfite-treated samples versus their respective untreated controls allows estimation of cytosine methylation percentage of the target, since methylated sequences will have high CG content compared to unmethylated targets after bisulfite treatment.
Compatible HRM Devices
Conducting HRM experiments requires compatible devices. Not all real-time PCR instruments can achieve the precise temperature increments necessary for HRM analysis. We recommend using one of the following qPCR thermocyclers from these manufacturers:
- Applied Biosystems: 7900, 7900HT, 7900HT FAST, StepOne, StepOne Plus, 7500, 7500 FAST, Viia7
- Bio-Rad: CFX96, CFX384
- BMS: Mic
- Eppendorf: Mastercycler ep realplex, Mastercycler realplex 2S
- Illumina: Eco
- Qiagen/Corbett: 6000, Q
- Roche Applied Science: Lightcycler 480, Lightcycler Nano
Other devices not listed above are also suitable for HRM analysis using the Clara® HRM Mix. Please use our qPCR Selection Tool, check the product guide of your device, or contact us at technical@pcrbio.com for assistance in selecting appropriate instrumentation.
Applications
- SNP Typing
- Gene scanning
- CpG methylation analysis
Technical Specifications
Clara® HRM Mix
| Components | 100 Reactions | 500 Reactions | 2000 Reactions | |
|---|---|---|---|---|
| 2x Clara HRM Mix | 1 x 1 mL | 5 x 1 mL | 20 x 1 mL |
Clara® HRM Mix
| Components | 100 Reactions | 500 Reactions | 2000 Reactions | |
|---|---|---|---|---|
| 2x Clara HRM Mix | 1 x 1 mL | 5 x 1 mL | 20 x 1 mL |
Reaction Information
| Reaction Volume | Storage | |||
|---|---|---|---|---|
| 20 μL |
Upon receipt, store the product between -30 °C and -20 °C. If stored correctly, the kit will retain full activity until the expiry date indicated. |
Instrument Compatibility
This product is compatible with all standard and fast qPCR instruments capable of High Resolution Melting. Use our qPCR Selection Tool to find compatible instruments.
Documents
Product Leaflet
User Manual
Safety Data Sheet
Application Note
Frequently Asked Questions (FAQs)
Can multiplexing be performed with Clara® HRM Mix?
Yes
How can unknown products be eliminated in HRM analysis?
Specificity can be enhanced by raising the annealing temperature or redesigning primers. Ensure no contaminating DNA as this could result in nonspecific signals. When setting up initial reactions, always run a gradient. We also recommend conducting agarose gel analysis for troubleshooting and product identification.
What are the general considerations to achieve high-quality results with Clara® HRM Mix?
- Ensure excellent DNA sample quality, meaning the DNA should be pure and free from inhibitors as this may affect melting temperatures. There should also be sufficient amounts of DNA in the sample.
- Follow standard precautions when setting up qPCR reactions, e.g., assembly on ice, precise pipetting, proper sealing of qPCR plates ensuring all wells have been sealed after cycling, spinning down samples to eliminate air bubbles and working clean.
- Your amplicon size should be in the range of 80 – 200 bp. A single base change will affect the melting temperature of a short sequence more than a long sequence. Larger amplicons can be analyzed but the resolution will be reduced. Conversely, very short amplicons will have low fluorescence and an unfavorable signal/noise ratio.
- Ensure your positive and negative controls are intact, not contaminated, and treated/purified in the same way as your samples to ensure consistency.
- Ensure your reaction reaches a plateau phase in each case. This will ensure a uniform starting fluorescence across the entire plate. Remember different product concentrations will melt slightly differently.
- Collect enough data points around the melting temperature of your amplicons.
- Examine your amplification plots. Poor amplification plots indicate issues in your qPCR and are sure to result in poor melting behavior.
What does Clara® HRM Mix contain?
Clara® HRM Mix is a ready-to-use mix. You only need to add primers, sample DNA, and PCR-grade water during reaction setup.
What should be the length of the amplification product?
To achieve efficient amplification under fast cycling conditions, we recommend the amplification product length of 80bp to 200bp. In all manufacturer's mixes, shorter amplicon lengths will allow the reaction to cycle faster. The length of amplification products should not exceed 400bp. Primers should have a predicted melting temperature of approximately 60°C, using default settings of Primer 3 (http://frodo.wi.mit.edu/primer3/).
Which devices are compatible with Clara® HRM Mix?
- Applied Biosystems: 7900, 7900HT, 7900HT FAST, StepOne, StepOne Plus, 7500, 7500 FAST, Viia7
- Bio-Rad: CFX96, CFX384
- BMS: Mic
- Eppendorf: Mastercycler ep realplex, Mastercycler realplex 2S
- Illumina: Eco
- Qiagen/Corbett: 6000, Q
- Roche Applied Science: Lightcycler 480, Lightcycler Nano
Why are unsatisfactory or discrepant results observed when comparing Clara® HRM Mix with a competitor's mix?
HRM mixes from different manufacturers can vary significantly in ionic strength and enhancement composition. Cycling conditions may work for one mix but may not be optimal for another; hence a cycling program that works with one mix might not apply to another without optimization. Cycling can be optimized by running a gradient for each amplicon, regardless of predicted or theoretical annealing temperatures.
If all of the above has been applied and results are still unsatisfactory, please email technical@pcrbio.com and include the following information:
- Amplicon size
- Reaction setup
- Cycling conditions
- Screenshots of amplification traces and melting profiles


Read more